CarbonTracker Lagrange Documentation
Spatial Distances¶
Calculate grid cell distances for use in calculating spatial covariances.
The calculation of the matrix Q in equation 2 depends on the spatial covariance matrix E. This matrix E is defined as 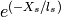 where
where 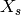 are the spatial distances between grid cells. These spatial distances need to be calculated only one time for a spatial domain. These distances are
calculated using the script make_sc.py:
are the spatial distances between grid cells. These spatial distances need to be calculated only one time for a spatial domain. These distances are
calculated using the script make_sc.py:
Usage: python make_sc.py [-c configfile]
where
configfile - configuration file. If not specified, use 'config.ini'.
Output:
A numpy save file with dimensions nlandcells x nlandcells, with distance in kilometers between each cell.
The name of this file is set in the configuration file with key 'sp_cov_file', e.g. sp_cov_file = "spcov.npy"
Example:
>>> import numpy
>>> a = numpy.load("spcov.npy")
>>> a
array([[ 0. , 109.46755851, 218.93484013, ...,
7671.1452984 , 7672.82817131, 7686.81558107],
[ 109.46755851, 0. , 109.46755851, ...,
7669.83574638, 7671.1452984 , 7683.27826395],
[ 218.93484013, 109.46755851, 0. , ...,
7668.90000321, 7669.83574638, 7680.10975617],
...,
[ 7671.1452984 , 7669.83574638, 7668.90000321, ...,
0. , 20.35208384, 122.06006263],
[ 7672.82817131, 7671.1452984 , 7669.83574638, ...,
20.35208384, 0. , 101.73045238],
[ 7686.81558107, 7683.27826395, 7680.10975617, ...,
122.06006263, 101.73045238, 0. ]])
>>> a.shape
(3470, 3470)
>>>
Spatial Correlation Length¶
Equation 2 uses a parameter ls for specifying the spatial covariance E. In the inversion code, a constant value of ls is used, and is defined in the configuration file with key name ‘spatial_corr_length’. The units for this value are in kilometers.
Example:
spatial_corr_length = 500.0
