CarbonTracker Lagrange Documentation
Stage 3 - Observations, background and priors¶
The solution of Equation 1 requires a prior estimate of s (called sp or sprior). The user must create the sprior file with dimensions ntimesteps x nlandcells, so there is a value for each land cell at each time step.
Equation 1 also uses z, a vector of measurements for use in the term
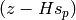 . This term is defined as
. This term is defined as
= (observation - bg - Hsp - Hother )
The program make_z.py makes these calculations and creates one file used in the inversion,
- zhsp.txt - contains
, dimensions (nobs)
and several additional files for information:
- obs-bg.txt - Observation values minus the background values.
- hsprior.txt - H convolved with sprior.
Prerequisites¶
- observation list - File with observation values, one value per line for each receptor.
- background list - File with background values, one value per line for each receptor.
- sprior - contains sp. Can be either a numpy save file, dimensions (ntimesteps x ncells), or a netcdf file, with a variable named ‘sprior’ and dimensions (ntimesteps x nlat x nlon)
- H blocks created by hsplit.py.
- (Optional) hother, one value per line for each receptor, with additional values to subtract from z.
Usage¶
make_z.py is run with:
Usage: make_z.py [--other=otherdata] [-c configfile] obsfile spriorfile backgroundfile where Reguired arguments obsfile : file with observation value, one line per receptor. spriorfile : file with sprior data. It can be either .npy or netcdf format. backgroundfile : file with background value, one line per receptor. Default is 'bg.txt'. Optional arguments -c configfile : file with configuration data. Default is 'config.ini'. --other=otherdata : file with other values to subtract from observations, one line per receptor. Output: zhsp.txt - Observations minus background minus hsprior minus hother hsprior.txt - H convolved with sprior obs-bg.txt - Observations minus background
